Experiments Confirm Sugary Shield Acts to Disguise SARS-CoV-2’s Spike
New results allow drug developers to understand areas of the spike most vulnerable to vaccines
October 6, 2020 | By Cynthia Dillon
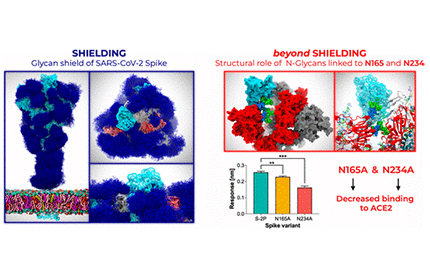 Amaro and her research colleagues used multiple microsecond-long, all-atom molecular dynamics simulations to provide an atomistic perspective on the roles of glycans and on the protein structure and dynamics of the virus’ shield, revealing an essential structural role of N-glycans at sites N165 and N234. Image by Lorenzo Casalino, Zied Gaieb (UC San Diego), et al.
Amaro and her research colleagues used multiple microsecond-long, all-atom molecular dynamics simulations to provide an atomistic perspective on the roles of glycans and on the protein structure and dynamics of the virus’ shield, revealing an essential structural role of N-glycans at sites N165 and N234. Image by Lorenzo Casalino, Zied Gaieb (UC San Diego), et al.
UC San Diego Department of Chemistry and Biochemistry’s Rommie Amaro, along with colleagues at Maynooth University in Ireland and the University of Texas at Austin (UT Austin), shared findings several weeks ago that revealed the atomic makeup of the sugary cloak that disguises SARS-CoV-2—the virus that can cause COVID-19—from the body’s immune system.
Their simulations and modeling demonstrated that molecules called glycans enable this disguise by changing the shape of the virus’ now familiar spike protein. An article posted recently by ACS Central Science describes their new experiments that validate those findings about the roles of glycans in the SARS-CoV-2 spike protein.
“We showed how clever the virus is, but now we have experimental proof that the glycan shield also acts as part of the ‘weaponry’ of the spike,” said Amaro, crediting Jason McLellan from UT Austin with the validating experiment.
According to Amaro, the new research results enable drug developers to understand what areas of the spike would be best to target with antibodies (vaccines), adding that they will generally want to target regions of the spike protein that are not hidden or masked from the human immune system by the glycan shield.
The researchers used the Texas Advanced Computing Center (TACC) Frontera supercomputer, with compute time made available through a Director’s Discretionary Allocation, made possible by the National Science Foundation (award no. OAC-1818253).

